14 Tree-Based Models
14.1 Overview of Models on Exam PA
GLM appears on every sitting since 2019.
Decision trees also appear on every version.
Regularized regression (lasso/ridge), bagging, random forests, GBMs appear less frequently (e.g., Dec 2019, Jun 2019).
Priority order:
1. GLM
2. Decision Trees
3. Boosting / Random Forests (when they appear)
Always include this in your executive summary:
After preparing features for modeling, I tested multiple models to identify factors affecting [target]. Each model was trained on 70% of the data (training set) and evaluated on the remaining 30% (test set). This split helps ensure models capture patterns and generalize to new data.
14.2 Bootstrap & Cross-Validation for Hyperparameter Tuning
Bootstrap = resampling with replacement to create many versions of the dataset.
Example: Predicting claim amounts
- Original data has observations A, B, C, D
- Bootstrap sample 1: A, A, B, D → different average claim
- Bootstrap sample 2: C, C, D, D → different average
Repeat hundreds of times → get distribution of parameter estimates (β₀, β₁, …) → compute means, confidence intervals empirically.
Cross-validation (more reliable):
- K-fold CV: Divide data into K parts
- Train on K-1 folds, test on 1 fold → repeat K times
- Average performance across folds
5-fold CV example:
Each fold gives slightly different model (different trees, different β values).
Average smooths out randomness.
14.3 Decision Trees – Core Idea
Goal: Partition feature space into regions with similar target values.
Example: Predict high-cost claim (> $20,000) using age and BMI.
- Start with root node
- Ask: Age < 20? → Yes/No branches
- Then: BMI < 15? → further split
- Each terminal (leaf) node gets majority class or average value
Terminology:
- Root: top node
- Branches: splits
- Depth/height: number of levels
- Leaf/Terminal nodes: final predictions
Practice exercise (from SRM sample):
Regression tree for log(claim amount) using age_cat and vehicle_age.
Trace three observations through the tree and report predicted values.
14.4 How Trees Choose Splits
Three criteria (only two commonly used on PA):
- Gini Index (most common)
- Range: 0 (perfect purity) to 1 (worst)
- Gini = 1 − Σ(pⱼ²) for each class j
- Weighted average across child nodes
- Range: 0 (perfect purity) to 1 (worst)
- Cross-Entropy / Information Gain
- Entropy = −Σ pⱼ log₂(pⱼ)
- Rarely appears on PA, but know definition
- Entropy = −Σ pⱼ log₂(pⱼ)
- Classification Error Rate (almost never used – too insensitive)
Example calculation (two possible splits):
- Split 1: Weighted Gini = 0.49
- Split 2: Weighted Gini = 0.39 → better split
14.5 Complexity Parameter & Pruning
complexity controls tree size (like λ in ridge/lasso).
Loss function:
SSE + complexity × (number of terminal nodes)
Pruning process:
- Grow large tree (small complexity)
- Look at complexity table / plot
- Choose complexity that balances error and complexity
- Cut branches with higher complexity
Bias-Variance Tradeoff:
- High complexity → simpler tree → high bias, low variance
- Low complexity → complex tree → low bias, high variance
14.6 Advantages & Disadvantages of Single Trees
Advantages:
- Easy to interpret
- Built-in variable selection
- Handles categorical variables without dummy encoding
- Captures non-linearities and interactions automatically
- Handles missing values (surrogate splits)
Disadvantages:
- Lower predictive accuracy than GLM / lasso / GBM / RF
- Predictions are piecewise constant (step function)
- High variance: small data change → very different tree
- Over-simplifies complex patterns
14.7 Bagging & Random Forests
Bagging = Bootstrap Aggregating
- Fit many trees on different bootstrap samples
- Average predictions (regression) or majority vote (classification)
- Reduces variance, keeps bias similar
Random Forest = Bagged trees + random feature subset at each split
- Extra randomness → more independent trees → lower variance
- Only one main tuning parameter: mtry (features per split)
Advantages:
- Resilient to overfitting
- Measures variable importance
- All single-tree benefits
Disadvantages:
- Harder to interpret
- Struggles with severe class imbalance (need stratified sampling / oversampling)
- Lower accuracy than boosting
- Cannot extrapolate beyond training range (regression)
14.8 Boosted Trees (GBM)
Boosting: sequential trees, each fits residuals of previous.
Process:
1. Start with mean(target)
2. Fit tree to residuals → get predictions
3. Update: new target = previous residuals − learning_rate × tree prediction
4. Repeat
Key parameters:
- Learning rate (shrinkage)
- Number of trees
- Tree depth / interaction.depth
- Subsample fraction
Advantages:
- Highest predictive accuracy on many problems
- Handles non-linearities, interactions, missing values, outliers
- Widely used in actuarial modeling and Kaggle winners
Disadvantages:
- Requires larger sample size
- Longer training time
- Easy to overfit → needs careful tuning / CV
14.9 Partial Dependence Plots
Show marginal effect of one (or two) predictors after averaging out others.
Useful for explaining: “How does age affect predicted charges, holding everything else constant?”
14.10 Final Exam Tips
- Know definitions: pruning, bagging, boosting, Gini, entropy, complexity
- Be ready to interpret tree output (splits, leaf values, % in each node)
- Compare models: interpretability vs accuracy
- Write concisely: “Random Forest averages many bagged trees to reduce variance. Boosting builds trees sequentially on residuals for higher accuracy but risks overfitting.”
For 2026 practice exams + solutions:
https://www.PredictiveInsightsAI.com
14.11 Load Dataset
datasets <- import("datasets")
ds <- datasets$load_dataset("supersam7/health_costs")
df <- as_tibble(ds$train$to_pandas())
glimpse(df)## Rows: 1,338
## Columns: 7
## $ age <dbl> 19, 18, 28, 33, 32, 31, 46, 37, 37, 60, 25, 62, 23, 56, 27, 1…
## $ sex <chr> "female", "male", "male", "male", "male", "female", "female",…
## $ bmi <dbl> 27.900, 33.770, 33.000, 22.705, 28.880, 25.740, 33.440, 27.74…
## $ children <dbl> 0, 1, 3, 0, 0, 0, 1, 3, 2, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0…
## $ smoker <chr> "yes", "no", "no", "no", "no", "no", "no", "no", "no", "no", …
## $ region <chr> "southwest", "southeast", "southeast", "northwest", "northwes…
## $ charges <dbl> 16884.924, 1725.552, 4449.462, 21984.471, 3866.855, 3756.622,…14.12 Single Decision Tree
tree_model <- rpart(charges ~ age + bmi + smoker + children + region,
data = df, method = "anova",
control = rpart.control(minsplit = 20, cp = 0.001))
rpart.plot(tree_model, type = 3, extra = 101,
main = "Decision Tree – Charges 2026")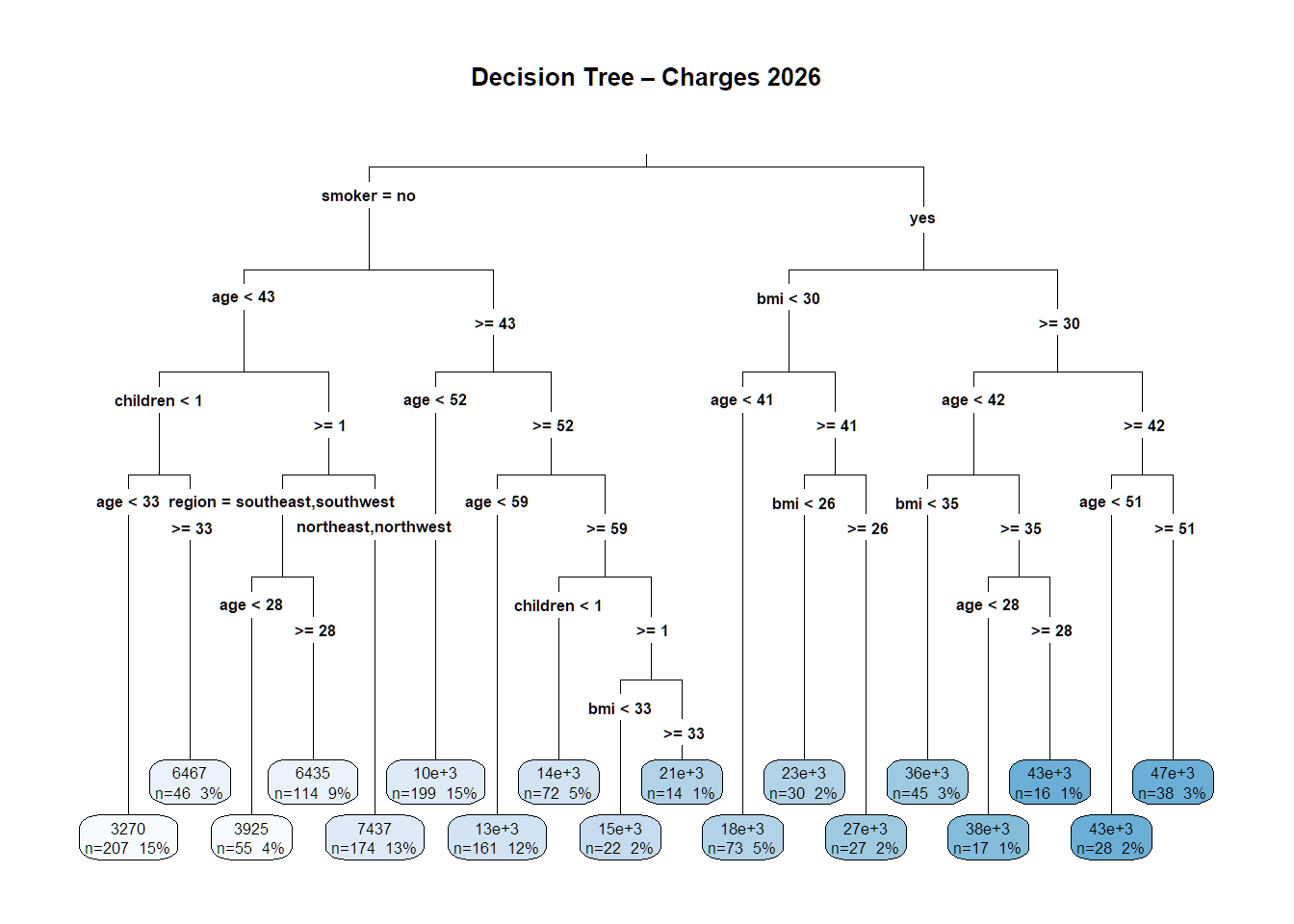
14.13 Random Forest
set.seed(42)
train_idx <- createDataPartition(df$charges, p = 0.8, list = FALSE)
train <- df[train_idx, ]
test <- df[-train_idx, ]
rf_control <- trainControl(method = "cv", number = 5)
rf_model <- train(charges ~ ., data = train,
method = "rf", trControl = rf_control,
tuneLength = 5, ntree = 100)
print(rf_model)## Random Forest
##
## 1072 samples
## 6 predictor
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 857, 858, 857, 858, 858
## Resampling results across tuning parameters:
##
## mtry RMSE Rsquared MAE
## 2 5220.551 0.8474560 3588.339
## 3 4557.605 0.8590567 2714.177
## 5 4509.468 0.8596150 2522.385
## 6 4532.766 0.8579675 2513.280
## 8 4602.214 0.8541628 2570.483
##
## RMSE was used to select the optimal model using the smallest value.
## The final value used for the model was mtry = 5.14.14 Gradient Boosting (gbm)
gbm_control <- trainControl(method = "cv", number = 5)
gbm_grid <- expand.grid(n.trees = c(100, 300),
interaction.depth = c(3, 5),
shrinkage = c(0.01, 0.1),
n.minobsinnode = c(10))
gbm_model <- train(charges ~ ., data = train,
method = "gbm", trControl = gbm_control,
tuneGrid = gbm_grid, verbose = FALSE)
print(gbm_model)## Stochastic Gradient Boosting
##
## 1072 samples
## 6 predictor
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 859, 857, 857, 858, 857
## Resampling results across tuning parameters:
##
## shrinkage interaction.depth n.trees RMSE Rsquared MAE
## 0.01 3 100 6283.666 0.8497361 4702.726
## 0.01 3 300 4460.758 0.8706725 2712.659
## 0.01 5 100 6088.864 0.8667520 4600.524
## 0.01 5 300 4350.976 0.8742149 2553.885
## 0.10 3 100 4329.199 0.8719095 2399.529
## 0.10 3 300 4519.625 0.8607597 2629.157
## 0.10 5 100 4398.602 0.8678141 2450.273
## 0.10 5 300 4650.457 0.8528952 2769.422
##
## Tuning parameter 'n.minobsinnode' was held constant at a value of 10
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were n.trees = 100, interaction.depth =
## 3, shrinkage = 0.1 and n.minobsinnode = 10.14.15 XGBoost
X_train <- model.matrix(charges ~ . -1, data = train)
X_test <- model.matrix(charges ~ . -1, data = test)
y_train <- train$charges
y_test <- test$charges
xgb_grid <- expand.grid(nrounds = c(200),
max_depth = c(3, 6),
eta = c(0.05, 0.1),
gamma = 0,
colsample_bytree = 0.8,
min_child_weight = 1,
subsample = 0.8)
xgb_model <- train(x = X_train, y = y_train,
method = "xgbTree",
trControl = trainControl(method = "cv", number = 5),
tuneGrid = xgb_grid,
verbosity = 0)
print(xgb_model)## eXtreme Gradient Boosting
##
## 1072 samples
## 9 predictor
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 857, 859, 857, 857, 858
## Resampling results across tuning parameters:
##
## eta max_depth RMSE Rsquared MAE
## 0.05 3 4390.945 0.8687443 2382.847
## 0.05 6 4777.860 0.8450791 2746.262
## 0.10 3 4540.453 0.8602340 2514.052
## 0.10 6 5023.069 0.8294898 3024.766
##
## Tuning parameter 'nrounds' was held constant at a value of 200
## Tuning
##
## Tuning parameter 'min_child_weight' was held constant at a value of 1
##
## Tuning parameter 'subsample' was held constant at a value of 0.8
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were nrounds = 200, max_depth = 3, eta
## = 0.05, gamma = 0, colsample_bytree = 0.8, min_child_weight = 1 and
## subsample = 0.8.14.16 Direct lightgbm
# Manual CV for LightGBM
dtrain <- lgb.Dataset(X_train, label = y_train)
set.seed(42)
folds <- createFolds(y_train, k=5)
params_list <- expand.grid(
max_depth = c(3,6,8),
num_leaves = c(20,31,50),
learning_rate = c(0.1)
)
rmse_results <- numeric(nrow(params_list))
for(i in 1:nrow(params_list)) {
params <- list(
objective = "regression",
metric = "rmse",
max_depth = params_list$max_depth[i],
num_leaves = params_list$num_leaves[i],
learning_rate = params_list$learning_rate[i],
feature_fraction = 0.8,
bagging_fraction = 0.8,
bagging_freq = 5,
verbose = -1
)
cv_rmse <- 0
for(fold_idx in folds) {
train_fold <- lgb.Dataset(X_train[-fold_idx,], label = y_train[-fold_idx])
val_fold <- lgb.Dataset(X_train[fold_idx,], label = y_train[fold_idx])
model <- lgb.train(params, train_fold, nrounds=200, valids=list(val=val_fold), early_stopping_rounds=20, verbose=0)
pred <- predict(model, X_train[fold_idx,])
cv_rmse <- cv_rmse + sqrt(mean((pred - y_train[fold_idx])^2))
}
rmse_results[i] <- cv_rmse / 5
}
# Best
best_i <- which.min(rmse_results)
best_params <- params_list[best_i,]
print(best_params) # Fix: use print
# Final train
lgb_model <- lgb.train(as.list(best_params), dtrain, nrounds=200, valids=list(test=dtest), early_stopping_rounds=20, verbose=0)
print(lgb_model)